Metabolic Engineering
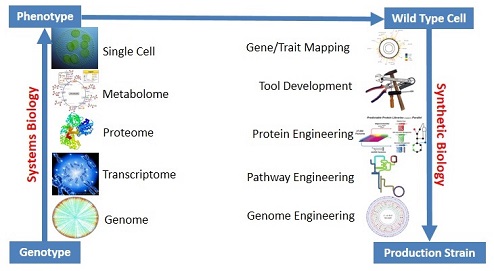
Metabolic engineering is altering genes and metabolic pathways in biological systems to produce molecules of interest. These molecules can be metabolites already found in the cell we are engineering, a metabolite found in another organism or a completely novel compound. In order to engineer cells to produce molecules of commercial relevance, we must leverage our knowledge of biological systems. We use modeling and –omics technologies to learn more about how the cell functions and how to overcome natural regulatory mechanisms. Advances in DNA synthesis and sequencing technologies have allowed us to learn more about the cells we are working with and design novel DNA sequences to introduce into the cells we are working with. The advent of synthetic biology and the push to standardize biological parts will also lead to faster strain design and development.
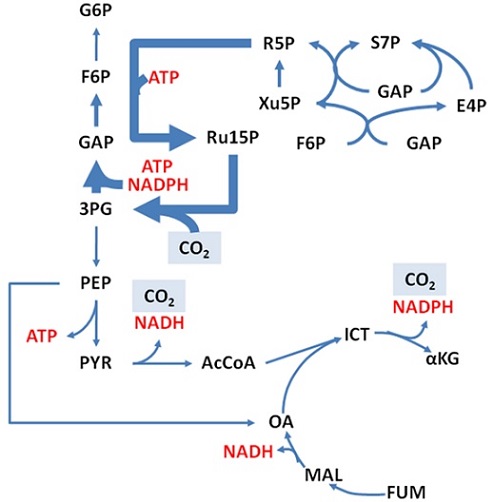
Metabolic engineers use metabolic models to create what is called a flux map (see below). In this flux map, the thickness of the arrows indicated the amount of carbon moving through each reaction. In this way, the flux map can be used like a traffic map – showing us where the most ‘traffic’ (or carbon) is and how to get from one metabolite to another in the most efficient way. These maps provide metabolic engineers with targets for genetic modification to improve yields.
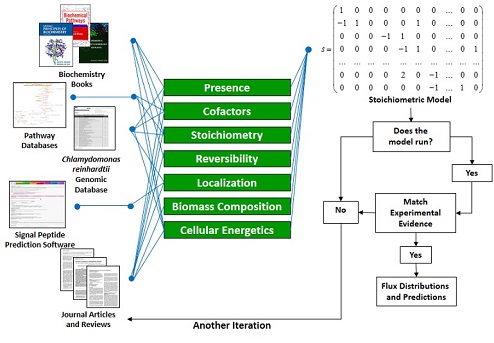
Metabolic models start with metabolic network reconstructions (see bleow). We collect information about all the enzymatic reactions occurring in the cell from a variety of sources. Then, we write mass balances on each metabolite in the cell based on the reaction stoichiometry. From there, we can build a model that can predict fluxes using optimization methods or calculate fluxes from experimental data.